|
Proposal
To study the molecular basis
for genetically ñ linked human birth defects.
The lab I intend to work in is at the Childrenís
Hospital of Philadelphia and is headed by a clinical
geneticist, Dr Ian Krantz. The goal of my summer internship
is to study the molecular basis for genetically ñ
linked human birth defects. Of particular interest are
developmental disorders caused by mutation of regulatory
genes. The identification of these genes and understanding
how the mutation of one or more of these genes can affect
many bodily systems is the all-important first step
in providing care to the patients at the hospital. Traditional
approaches such as chromosomal rearrangements and linkage
analysis as well as less conventional ones are used
to map these genes of interest. This will lead to a
better understanding of the molecular basis of human
embryonic development and the defects therein. Several
developmental disorders are being studied in this context,
including Cornelia de Lange syndrome, hearing loss,
Alagille syndrome and congenital heart defects. Over
the course of ten weeks during this summer I will be
working closely with medical students, post doctoral
students and other undergraduates on many of these projects
and attending seminars on other researchersí
work at the Childrenís Hospital and at the University
of Pennsylvania.
Summary
Candidate genes for non-syndromic congenital
hearing loss
This summer I worked in a genetics lab
that studies the molecular basis of genetically-linked
human developmental disorders. Research has been conducted
on Alagille Syndrome, Cornelia de Lange syndrome, hearing
loss and congenital heart defects. For a great deal
of the summer I focused on the identification of novel
genes for hearing loss. Hearing loss is interesting
in that many genes have been implicated in the disorder
and as many as 70% of hearing loss cases seem to have
a genetic cause. The project focused on non-syndromic
hearing loss, which has no other symptoms or structural
defects associated with the hearing loss. The etiology
of non-syndromic hearing loss is not well characterized,
but it is a highly genetically heterogeneous diagnosis
with approximately 100 genetic loci implicated. Identification
of small genomic rearrangements using new technology
would help in identifying causative genes and new loci
for hearing loss.
The first step in this project was genome-wide SNP genotyping
of over 600 bilateral sensorineural hearing loss probands.
Copy number variations, deletions and duplications in
the genome, were called by an algorithm which lists
all of the copy number variations (CNVs) in a useable
format. We use another computer-based algorithm developed
in our lab to screen through these lists and to rank
the CNVs based on their likelihood to be pathogenic.
New techniques allow us to see smaller changes in the
genome than before, which is very important for our
purposes. After researching the genes involved in the
highest ranked CNVs a few candidates are selected for
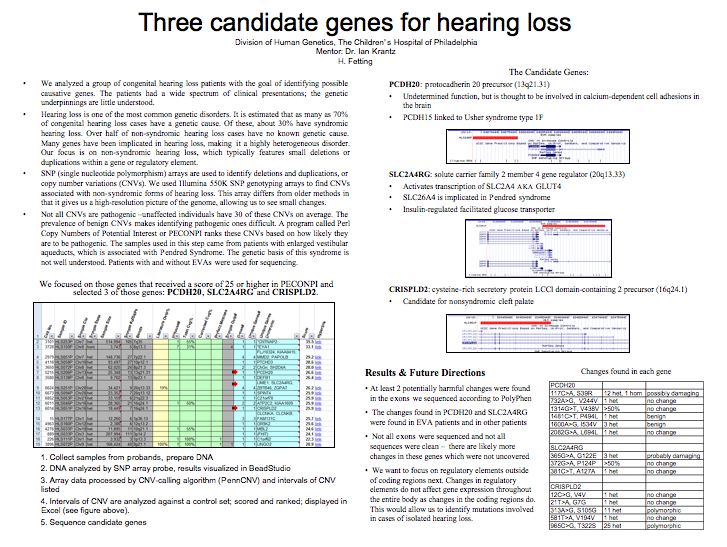
sequencing. We chose PCDH20, a protocadherin precursor
thought to be involved in calcium-dependent cell adhesions
in the brain; SLC2A4RG, which regulates GLUT4; and CRISPLD2.
All of the CNVs involved deletions in exonic regions,
were relatively small and were not covered by literature
or in-house controls. As of this writing, sequencing
for these genes has not been completed. At least two
potentially deleterious changes were found, one in PCDH20
(117C>A, S39R) and one in SLC2A4RG (365G>A, G122E).
Further characterization needs to be done before validating
these changes.
Future work will focus more on regulatory elements lying
outside of the coding regions. Changes in these regulatory
elements are more likely to be implicated in many non-syndromic
hearing loss cases than large exonic changes. Large
changes in coding regions can affect gene expression
in many cell types throughout the body, whereas changes
in regulatory elements affect only the genes they regulate.
These changes are more specific and may account for
many cases of isolated hearing loss. Changes to the
scoring parameters of our CNV ranking program must be
made in order to identify and accordingly score these
important regions. The next project will focus on using
this approach with samples from patients with congenital
heart defects.
Poster
(saved as a pdf)
|

